bash
docker run -it ghcr.io/js2264/biocbookdemo:devel R
Package: BiocBookDemo
Authors: Jacques Serizay [aut, cre]
Compiled: 2023-12-08
Package version: 1.1.3
R version: R Under development (unstable) (2023-11-22 r85609)
BioC version: 3.19
License: MIT + file LICENSE
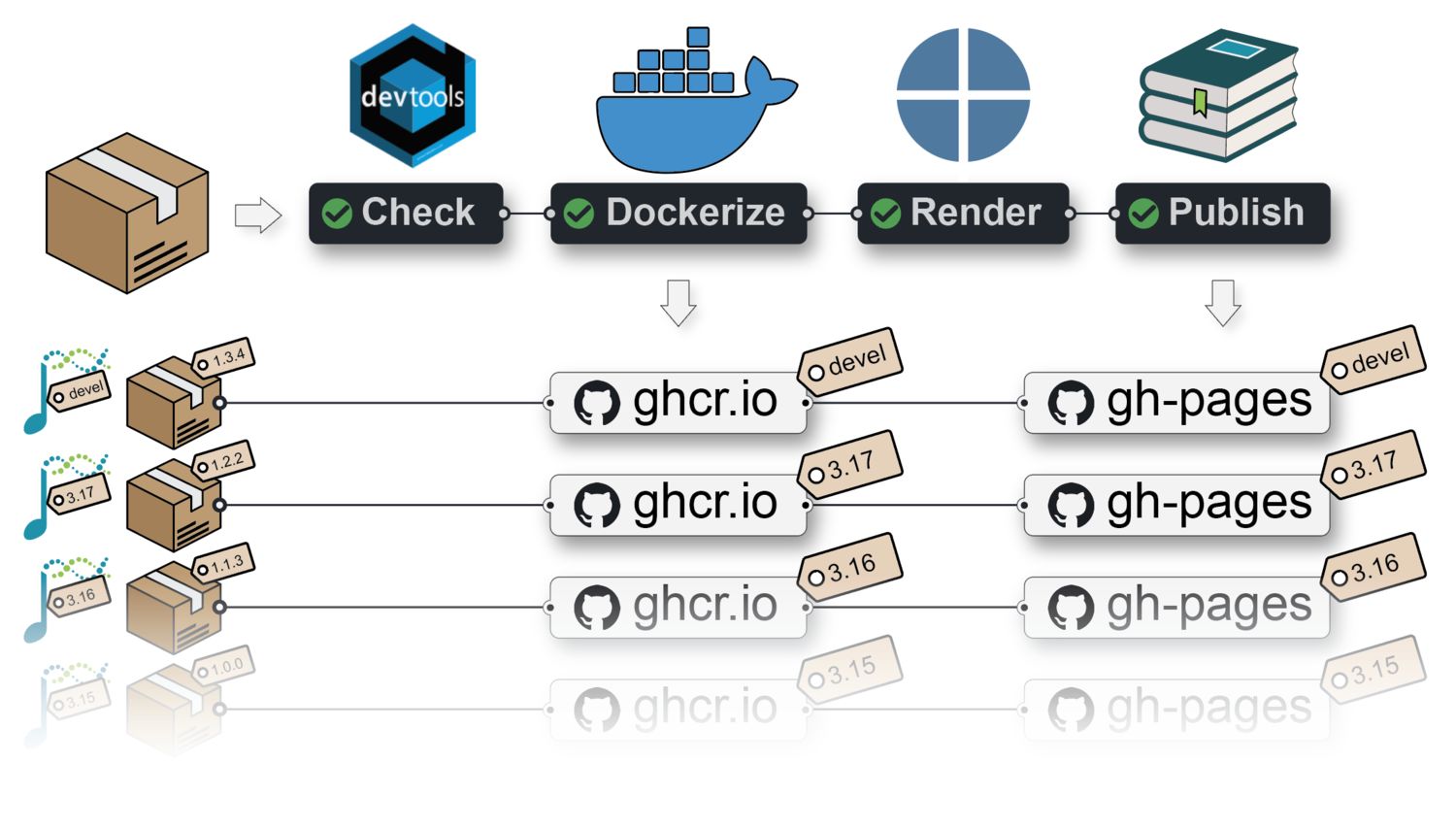
BiocBooks?BiocBooks are package-based, versioned online books with a supporting Docker image for each book version.
A BiocBook can be created by authors (e.g. R developers, but also scientists, teachers, communicators, …) who wish to:
A {BiocBook}-based package hosted on GitHub with a branch named RELEASE_X_Y provides:
gh-pages branch;Both are built against the specific Bioconductor release X.Y.
A {BiocBook}-based package submitted to Bioconductor also lead to the online book being independently built by the Bioconductor Build System (BBS) and deployed to https://bioconductor.org/books/<bioc_version>/<pkg>/
BiocBook} package?The {BiocBook} package offers a streamlined approach to creating BiocBooks, with several important benefits:
BiocBook}-based package without leaving R;pages/*.qmd files using enhanced markdown;BiocBook}-based package to Bioconductor.The containerization and publishing of the new {BiocBook}-based package is automated:
BiocBooksWhen a {BiocBook}-based package is accepted into Bioconductor, it is automatically integrated into the Bionconductor Build System (BBS).
This means that it is getting built using R CMD build --keep-empty-dirs --no-resave-data .. This triggers the rendering of the book contained in /inst/. Book packages built by the BBS are then automatically deployed and are eventually available at https://bioconductor.org/books/<bioc_version>/<pkg>/.
A separate Docker image is built for each branch (named devel or RELEASE_X_Y) of a {BiocBook}-based Github repository.
Each Docker image provides pre-installed R packages:
X.Y;X.Y (listed in DESCRIPTION);The Docker images also include a micromamba-based environment, named BiocBook, in which all the softwares listed in requirements.yml are installed.
For example, Docker images built from the {BiocBookDemo} package repository are available here:
👉 ghcr.io/js2264/biocbookdemo 🐳
You can get access to all the packages used in this book in < 1 minute, using this command in a terminal:
bash
docker run -it ghcr.io/js2264/biocbookdemo:devel RRegardless of whether the book package is submitted to Bioconductor, a Github Actions workflow publishes individual online books for each branch (named devel or RELEASE_X_Y) of a BiocBook-based Github repository.
For example, the online book version matching the devel version of the {BiocBook} package is available from:
An RStudio Server instance based on a specific Bioconductor <version> (devel or RELEASE_X_Y) can be initiated from the corresponding Docker image as follows:
bash
docker run \
--volume <local_folder>:<destination_folder> \
-e PASSWORD=OHCA \
-p 8787:8787 \
ghcr.io/<github_user>/<biocbook_repo>:<version>The initiated RStudio Server instance will be available at https://localhost:8787.
Further instructions regarding Bioconductor-based Docker images are available here.
This works was inspired by and closely follows the strategy used in coordination by the Bioconductor core team and Aaron Lun to submit book-containing packages (from the OSCA series as well as SingleR and csaw books).
This package was also inspired by the *down package series, including:
sessioninfo::session_info(
installed.packages()[,"Package"],
include_base = TRUE
)
## ─ Session info ────────────────────────────────────────────────────────────
## setting value
## version R Under development (unstable) (2023-11-22 r85609)
## os Ubuntu 22.04.3 LTS
## system x86_64, linux-gnu
## ui X11
## language (EN)
## collate C
## ctype en_US.UTF-8
## tz Etc/UTC
## date 2023-12-08
## pandoc 3.1.1 @ /usr/local/bin/ (via rmarkdown)
##
## ─ Packages ────────────────────────────────────────────────────────────────
## package * version date (UTC) lib source
## askpass 1.2.0 2023-09-03 [2] CRAN (R 4.4.0)
## base * 4.4.0 2023-11-26 [3] local
## base64enc 0.1-3 2015-07-28 [2] CRAN (R 4.4.0)
## BiocBookDemo 1.1.3 2023-12-08 [1] local
## BiocManager 1.30.22 2023-08-08 [2] CRAN (R 4.4.0)
## BiocStyle 2.31.0 2023-10-24 [2] Bioconductor
## BiocVersion 3.19.1 2023-10-26 [2] Bioconductor
## bookdown 0.37 2023-12-01 [2] CRAN (R 4.4.0)
## boot 1.3-28.1 2022-11-22 [3] CRAN (R 4.4.0)
## brew 1.0-8 2022-09-29 [2] CRAN (R 4.4.0)
## brio 1.1.3 2021-11-30 [2] CRAN (R 4.4.0)
## bslib 0.6.1 2023-11-28 [2] CRAN (R 4.4.0)
## cachem 1.0.8 2023-05-01 [2] CRAN (R 4.4.0)
## callr 3.7.3 2022-11-02 [2] CRAN (R 4.4.0)
## class 7.3-22 2023-05-03 [3] CRAN (R 4.4.0)
## cli 3.6.1 2023-03-23 [2] CRAN (R 4.4.0)
## clipr 0.8.0 2022-02-22 [2] CRAN (R 4.4.0)
## cluster 2.1.5 2023-11-27 [3] CRAN (R 4.4.0)
## codetools 0.2-19 2023-02-01 [3] CRAN (R 4.4.0)
## commonmark 1.9.0 2023-03-17 [2] CRAN (R 4.4.0)
## compiler 4.4.0 2023-11-26 [3] local
## cpp11 0.4.6 2023-08-10 [2] CRAN (R 4.4.0)
## crayon 1.5.2 2022-09-29 [2] CRAN (R 4.4.0)
## credentials 2.0.1 2023-09-06 [2] CRAN (R 4.4.0)
## curl 5.1.0 2023-10-02 [2] CRAN (R 4.4.0)
## datasets * 4.4.0 2023-11-26 [3] local
## desc 1.4.2 2022-09-08 [2] CRAN (R 4.4.0)
## devtools 2.4.5 2022-10-11 [2] CRAN (R 4.4.0)
## diffobj 0.3.5 2021-10-05 [2] CRAN (R 4.4.0)
## digest 0.6.33 2023-07-07 [2] CRAN (R 4.4.0)
## docopt 0.7.1 2020-06-24 [2] CRAN (R 4.4.0)
## downlit 0.4.3 2023-06-29 [2] CRAN (R 4.4.0)
## ellipsis 0.3.2 2021-04-29 [2] CRAN (R 4.4.0)
## evaluate 0.23 2023-11-01 [2] CRAN (R 4.4.0)
## fansi 1.0.5 2023-10-08 [2] CRAN (R 4.4.0)
## fastmap 1.1.1 2023-02-24 [2] CRAN (R 4.4.0)
## fontawesome 0.5.2 2023-08-19 [2] CRAN (R 4.4.0)
## foreign 0.8-86 2023-11-28 [3] CRAN (R 4.4.0)
## fs 1.6.3 2023-07-20 [2] CRAN (R 4.4.0)
## gert 2.0.0 2023-09-26 [2] CRAN (R 4.4.0)
## gh 1.4.0 2023-02-22 [2] CRAN (R 4.4.0)
## gitcreds 0.1.2 2022-09-08 [2] CRAN (R 4.4.0)
## glue 1.6.2 2022-02-24 [2] CRAN (R 4.4.0)
## graphics * 4.4.0 2023-11-26 [3] local
## grDevices * 4.4.0 2023-11-26 [3] local
## grid 4.4.0 2023-11-26 [3] local
## highr 0.10 2022-12-22 [2] CRAN (R 4.4.0)
## htmltools 0.5.7 2023-11-03 [2] CRAN (R 4.4.0)
## htmlwidgets 1.6.3 2023-11-22 [2] CRAN (R 4.4.0)
## httpuv 1.6.12 2023-10-23 [2] CRAN (R 4.4.0)
## httr 1.4.7 2023-08-15 [2] CRAN (R 4.4.0)
## httr2 1.0.0 2023-11-14 [2] CRAN (R 4.4.0)
## ini 0.3.1 2018-05-20 [2] CRAN (R 4.4.0)
## jquerylib 0.1.4 2021-04-26 [2] CRAN (R 4.4.0)
## jsonlite 1.8.8 2023-12-04 [2] CRAN (R 4.4.0)
## KernSmooth 2.23-22 2023-07-10 [3] CRAN (R 4.4.0)
## knitr 1.45 2023-10-30 [2] CRAN (R 4.4.0)
## later 1.3.1 2023-05-02 [2] CRAN (R 4.4.0)
## lattice 0.22-5 2023-10-24 [3] CRAN (R 4.4.0)
## lifecycle 1.0.4 2023-11-07 [2] CRAN (R 4.4.0)
## littler 0.3.18 2023-03-26 [2] CRAN (R 4.4.0)
## magrittr 2.0.3 2022-03-30 [2] CRAN (R 4.4.0)
## MASS 7.3-60.1 2023-11-26 [3] local
## Matrix 1.6-4 2023-11-30 [3] CRAN (R 4.4.0)
## memoise 2.0.1 2021-11-26 [2] CRAN (R 4.4.0)
## methods * 4.4.0 2023-11-26 [3] local
## mgcv 1.9-0 2023-07-11 [3] CRAN (R 4.4.0)
## mime 0.12 2021-09-28 [2] CRAN (R 4.4.0)
## miniUI 0.1.1.1 2018-05-18 [2] CRAN (R 4.4.0)
## nlme 3.1-164 2023-11-27 [3] CRAN (R 4.4.0)
## nnet 7.3-19 2023-05-03 [3] CRAN (R 4.4.0)
## openssl 2.1.1 2023-09-25 [2] CRAN (R 4.4.0)
## parallel 4.4.0 2023-11-26 [3] local
## pillar 1.9.0 2023-03-22 [2] CRAN (R 4.4.0)
## pkgbuild 1.4.2 2023-06-26 [2] CRAN (R 4.4.0)
## pkgconfig 2.0.3 2019-09-22 [2] CRAN (R 4.4.0)
## pkgdown 2.0.7 2022-12-14 [2] CRAN (R 4.4.0)
## pkgload 1.3.3 2023-09-22 [2] CRAN (R 4.4.0)
## praise 1.0.0 2015-08-11 [2] CRAN (R 4.4.0)
## preprocessCore 1.65.0 2023-10-24 [2] Bioconductor
## prettyunits 1.2.0 2023-09-24 [2] CRAN (R 4.4.0)
## processx 3.8.2 2023-06-30 [2] CRAN (R 4.4.0)
## profvis 0.3.8 2023-05-02 [2] CRAN (R 4.4.0)
## promises 1.2.1 2023-08-10 [2] CRAN (R 4.4.0)
## ps 1.7.5 2023-04-18 [2] CRAN (R 4.4.0)
## purrr 1.0.2 2023-08-10 [2] CRAN (R 4.4.0)
## R6 2.5.1 2021-08-19 [2] CRAN (R 4.4.0)
## ragg 1.2.6 2023-10-10 [2] CRAN (R 4.4.0)
## rappdirs 0.3.3 2021-01-31 [2] CRAN (R 4.4.0)
## rcmdcheck 1.4.0 2021-09-27 [2] CRAN (R 4.4.0)
## Rcpp 1.0.11 2023-07-06 [2] CRAN (R 4.4.0)
## rematch2 2.1.2 2020-05-01 [2] CRAN (R 4.4.0)
## remotes 2.4.2.1 2023-07-18 [2] CRAN (R 4.4.0)
## rlang 1.1.2 2023-11-04 [2] CRAN (R 4.4.0)
## rmarkdown 2.25 2023-09-18 [2] CRAN (R 4.4.0)
## roxygen2 7.2.3 2022-12-08 [2] CRAN (R 4.4.0)
## rpart 4.1.21 2023-10-09 [3] CRAN (R 4.4.0)
## rprojroot 2.0.4 2023-11-05 [2] CRAN (R 4.4.0)
## rstudioapi 0.15.0 2023-07-07 [2] CRAN (R 4.4.0)
## rversions 2.1.2 2022-08-31 [2] CRAN (R 4.4.0)
## sass 0.4.8 2023-12-06 [2] CRAN (R 4.4.0)
## sessioninfo 1.2.2 2021-12-06 [2] CRAN (R 4.4.0)
## shiny 1.8.0 2023-11-17 [2] CRAN (R 4.4.0)
## sourcetools 0.1.7-1 2023-02-01 [2] CRAN (R 4.4.0)
## spatial 7.3-17 2023-07-20 [3] CRAN (R 4.4.0)
## splines 4.4.0 2023-11-26 [3] local
## stats * 4.4.0 2023-11-26 [3] local
## stats4 4.4.0 2023-11-26 [3] local
## stringi 1.8.2 2023-11-23 [2] CRAN (R 4.4.0)
## stringr 1.5.1 2023-11-14 [2] CRAN (R 4.4.0)
## survival 3.5-7 2023-08-14 [3] CRAN (R 4.4.0)
## sys 3.4.2 2023-05-23 [2] CRAN (R 4.4.0)
## systemfonts 1.0.5 2023-10-09 [2] CRAN (R 4.4.0)
## tcltk 4.4.0 2023-11-26 [3] local
## testthat 3.2.0 2023-10-06 [2] CRAN (R 4.4.0)
## textshaping 0.3.7 2023-10-09 [2] CRAN (R 4.4.0)
## tibble 3.2.1 2023-03-20 [2] CRAN (R 4.4.0)
## tinytex 0.49 2023-11-22 [2] CRAN (R 4.4.0)
## tools 4.4.0 2023-11-26 [3] local
## urlchecker 1.0.1 2021-11-30 [2] CRAN (R 4.4.0)
## usethis 2.2.2 2023-07-06 [2] CRAN (R 4.4.0)
## utf8 1.2.4 2023-10-22 [2] CRAN (R 4.4.0)
## utils * 4.4.0 2023-11-26 [3] local
## vctrs 0.6.5 2023-12-01 [2] CRAN (R 4.4.0)
## waldo 0.5.2 2023-11-02 [2] CRAN (R 4.4.0)
## whisker 0.4.1 2022-12-05 [2] CRAN (R 4.4.0)
## withr 2.5.2 2023-10-30 [2] CRAN (R 4.4.0)
## xfun 0.41 2023-11-01 [2] CRAN (R 4.4.0)
## xml2 1.3.5 2023-07-06 [2] CRAN (R 4.4.0)
## xopen 1.0.0 2018-09-17 [2] CRAN (R 4.4.0)
## xtable 1.8-4 2019-04-21 [2] CRAN (R 4.4.0)
## yaml 2.3.7 2023-01-23 [2] CRAN (R 4.4.0)
## zip 2.3.0 2023-04-17 [2] CRAN (R 4.4.0)
##
## [1] /tmp/Rtmpdf9t5c/Rinst55b4648fc
## [2] /usr/local/lib/R/site-library
## [3] /usr/local/lib/R/library
##
## ───────────────────────────────────────────────────────────────────────────